Drug-TTA: Test-Time Adaptation for Drug Virtual Screening via Multi-task Meta Auxiliary Learning
Ao Shen*, Mingzhi Yuan*, Yingfan Ma, Jie Du, Qiao Huang, Manning Wang☨
The Forty-Second International Conference on Machine Learning
Abstract
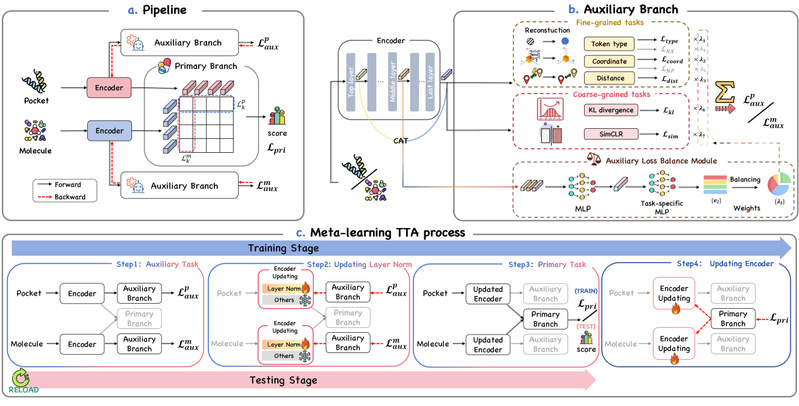
Virtual screening is a critical step in drug discovery, aiming at identifying potential drugs that bind to a specific protein pocket from a large database of molecules. Traditional docking methods are time-consuming, while learning-based approaches supervised by high-precision conformational or affinity labels are limited by the scarcity of training data. Recently, a paradigm of feature alignment through contrastive learning has gained widespread attention. This method does not require explicit binding affinity scores, but it suffers from the issue of overly simplistic construction of negative samples, which limits their generalization to more difficult test cases. In this paper, we propose Drug-TTA, which leverages a large number of self-supervised auxiliary tasks to adapt the model to each test instance. Specifically, we incorporate the auxiliary tasks into both the training and the inference process via meta-learning to improve the performance of the primary task of virtual screening. Additionally, we design a multi-scale feature based Auxiliary Loss Balance Module (ALBM) to balance the auxiliary tasks to improve their efficiency. Extensive experiments demonstrate that Drug-TTA achieves state-of-the-art (SOTA) performance in all five virtual screening tasks under a zero-shot setting, showing an average improvement of 9.86% in AUROC metric compared to the baseline without test-time adaptation.