White Matter Biomarker for Predicting De Novo Parkinson's Disease Using Tract-Based Spatial Statistics: A Machine Learning-Based Model
Qi Zhang, Haoran Wang, Yonghong Shi†, and Wensheng Li†
Quantitative Imaging in Medicine and Surgery (IF = 2.8)
Abstract
Background: Parkinson's disease (PD) is an irreversible, chronic degenerative disease of the central nervous system, potentially associated with cerebral white matter (WM) lesions. Investigating the microstructural alterations within the WM in the early stages of PD can help to identify the disease early and enable intervention to reduce the associated serious threats to health.
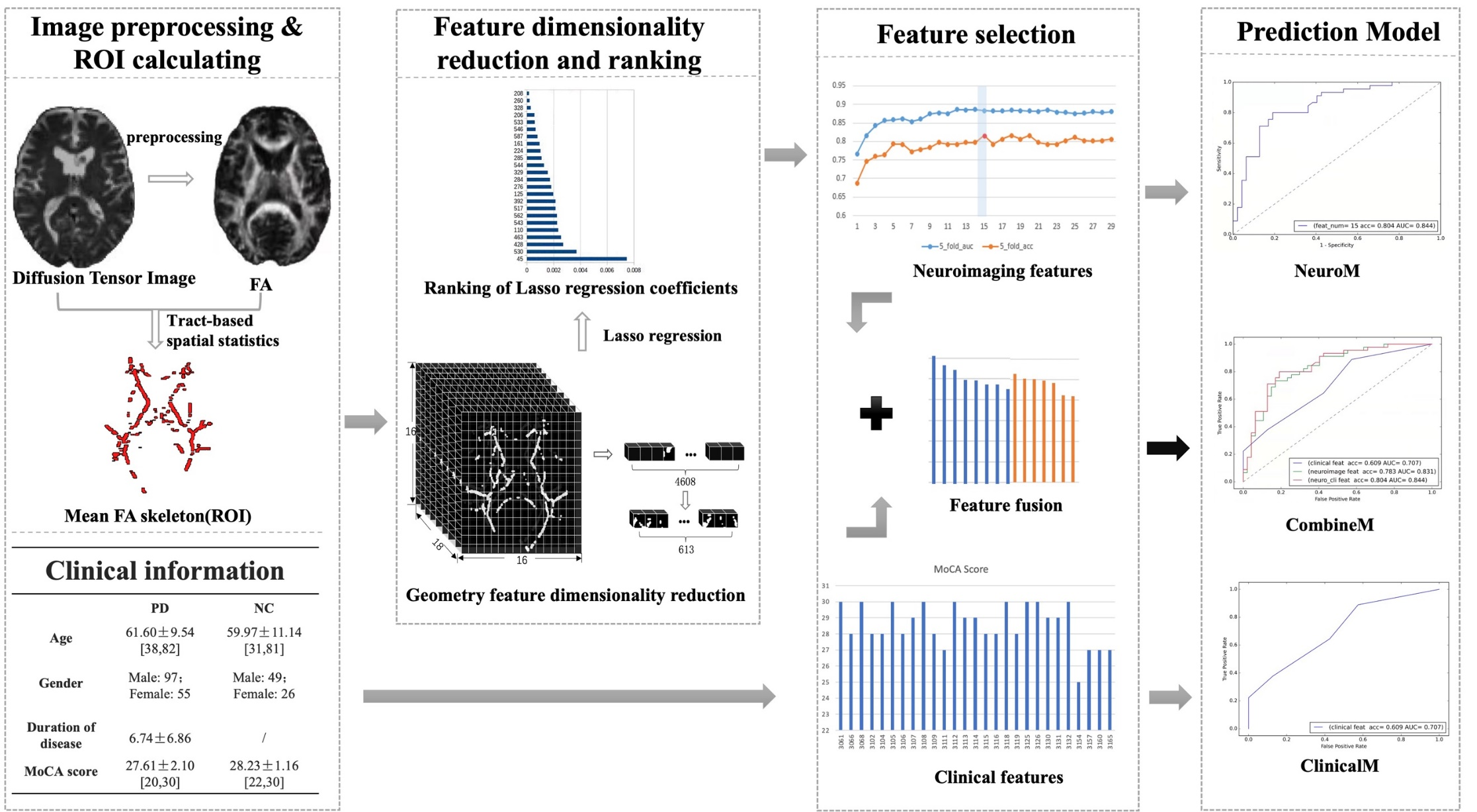
Methods: This study selected 227 cases from the Parkinson's Progression Markers Initiative (PPMI) database, including 152 de novo PD patients (PDs) and 75 normal controls (NC). Whole-brain voxel analysis of the WM was performed using the tract-based spatial statistics (TBSS) method. The WM regions with statistically significant differences (P<0.05) between the PD and NC groups were identified and used as masks. The mask was applied to each case’s fractional anisotropy (FA) image to extract voxel values as feature vectors. Geometric dimensionality reduction was then applied to eliminate redundant values in the feature vectors. Subsequently, the cases were randomly divided into a training group (158 cases, including 103 PDs and 55 NC) and a test group (69 cases, including 49 PDs and 20 NC). The least absolute shrinkage and selection operator (LASSO) regression algorithm was employed to extract the minimal set of relevant features, then the random forest (RF) algorithm was utilized for classification using 5-fold cross validation. The resulting model was further integrated with clinical factors to create a comprehensive prediction model.
Results: In comparison to the NC group, the FA values in PDs exhibited a statistically significant decrease (P<0.05), indicating the presence of widespread WM lesions across multiple brain regions. Moreover, the PD prediction model, constructed based on these WM lesion regions, yielded prediction accuracy (ACC) and area under the receiver operating characteristic (ROC) curve (AUC) values of 0.778 and 0.865 in the validation set, and 0.783 and 0.831 in the test set, respectively. Furthermore, the performance of the integrated model showed some improvement, with ACC and AUC values in the test set reaching 0.804 and 0.844, respectively.
Conclusions: The quantitative calculation of WM lesion area on FA images using the TBSS method can serve as a neuroimaging biomarker for diagnosing and predicting early PD at the individual level. When integrated with clinical variables, the predictive performance improves.